_
|
|
|||||||||||||||||||||||
|
Evaluation
of the Invader® assay
platform for the
detection of allelic variants:
|
||||||||||||||||||||||||
|
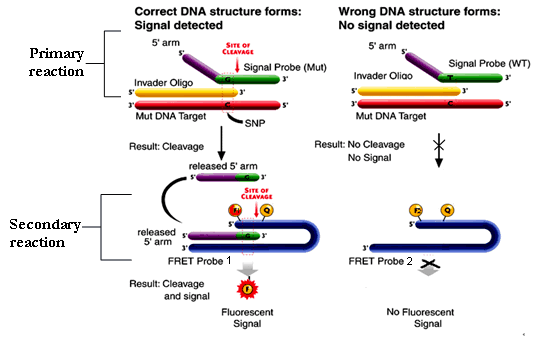
Project Leader: Dr Dave Bunyan Company website: www.twt.com (Third Wave™ Technologies) Invader® assay technology: The Invader® DNA assay enables simultaneous detection of
two different alleles in a bi-allelic system in a non-PCR based
assay. Two oligonucleotide probes (an allele specific primary
probe and an Invader® probe) hybridise in tandem to the target
DNA to form a specific overlapping structure. The 5’-end
of the primary probe contains a 5’-Flap that is non-complementary
to the target DNA and so is unable to hybridise to the target
sequence. The 3’-end of the bound Invader® probe overlaps
the primary probe by a single base this is the site of the allelic
variant or SNP. The target specific 5’-Flap oligos’s are involved in a secondary reaction where they act as Invader® probes on a Fluorescent resonance energy transfer (FRET™) cassette leading to the formation of an overlapping structure that is recognised by the Cleavase® enzyme. When the FRET™ cassette is cleaved a fluorophore is released form a quencher on the FRET™ generating a fluorescence signal. There are two signal fluorophores attached to two different FRET™ cassettes (FRET™ 1 and FRET™ 2) that are spectrally distinct and specific to either allele of the bi-allelic system. The ratios of the two fluorescent signals then allows a genotype to be assigned.
The reaction conditions (63oC) allow cycling of the primary probes and Invader® probes meaning multiple rounds of primary probe cleavage occurs per DNA target and multiple fluorescent signals are generated per DNA target.
The Invader® assay was evaluated in conjunction with other SNP detection technologies (Pyrosequencing™ and NanoChip®) by testing 100 anonymised thrombophilia patients samples for the Factor V Leiden (G1691A), Factor II (G20210A ) & MTHFR (C677T) variants that had previously been genotyped using restriction fragment length polymorphism (PCR based assay). The FVL and FII results display 100% concordance to the RFLP method and testing of the MTHFR variant is currently being undertaken. Evaluation of the Invader® assay platform for molecular analysis of the Factor V (G1691A) and Factor II (G20210A) mutations. Presentation to the CMGS Spring Meeting in 2004 available for download here.
|
||||||||||||||||||||||||
Last Updated: 6 August, 2008 by G.Watkins |
||||||||||||||||||||||||