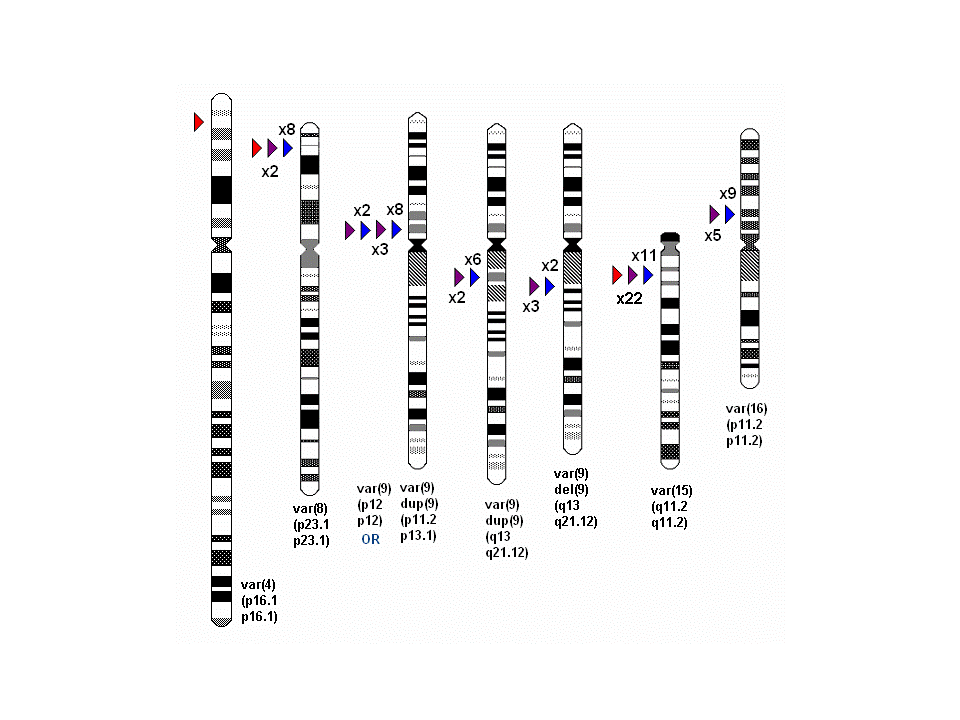
Euchromatic variants (EVs) - large scale cytogenetically visible copy number variants
Band
var(4)
(p16.1p16.1)
var(8)
(p23.1p23.1)
var(9)
(p12p12)
var(9)dup(9)
(p11.2p13.1)
var(9)dup(9) OR
var(9)trp(9)
(q13q21.12))
var(9)
del(9) OR
var(9) dup(9)
(q13q21.12)
var(15)
(q11.2q11.2)
var(16)
(p11.2p11.2)
Content
DEFB/OR/
BUB and
DRD5 genes
DEFB/OR
genes/
pseudo-
genes
1.0 Mb
repeat
Multiple genes, pseudo-genes, gene fragments and CNVs
NF1/IGVH/
GABRA5/
BCL8A
pseudo/genes
IGVH/CTD/
ADL/
FLJ46121/
TP53TG3
pseudo/genes
Control
copy number
2
2 - 7
?4
4
4
4
NF1: 1-4
IGVH: 1-3
4
Variant
copy number
6
9 - 12
7 - 14
5 - 6
5 OR 6
3 OR 5
NF1: 5-10
IGVH: 4-9
10 - 12
Balikova
et al
Am J Hum Genet
2003;82:
181-187
Hollox et al
Am J Hum Genet
2003;73:
591-600
Lecce et al
Hum Genet
2006;118:
760-766
Di Giacomo
et al
Prenat Diagn, 2004;24:619-622
Willatt et al
Eur J Hum Genet, 2007;15:45-52 Mevatee et al
Chromosome Res 13 Suppl 1 1.33P
Fantes et al
J Med Genet
2002;39:170-177
Barber et al
Hum Genet
1999;104:211-218
Euchromatic Variant (EV) Chart
Idiograms with the position at which EVs occur marked by arrows.
Group 1 EV imbalances are in blue; Group 2 EV in purple and Group 3 EV in red.
Figures give the number of times independent families with the same rearrangement have been reported (e.g. 8 etc).
| Submission Form |
National Genetics Reference Laboratory
(Wessex)
Salisbury District Hospital
Salisbury